We develop methods and tools for information retrieval, knowledge representation, model inference, and analysis of complex systems. Our goal is to understand and explain the behavior of these systems, predict their responses to external influences or internal perturbations, and recommend new interventions and designs. Leveraging advancements in neuro-symbolic artificial intelligence, we aim to understand, explain, and design complex dynamic systems in the presence of uncertainty, perturbations, and conflicting information. We study biological systems such as immune system, cancer microenvironment, and infectious diseases, as well as combined social, economic, political, and agricultural world systems. Our interdisciplinary lab includes members with backgrounds in electrical and computer engineering, bioengineering, mechanical engineering, computer science, computational biology, physics, and biology.
DySE - Dynamic Systems Explanation Framework
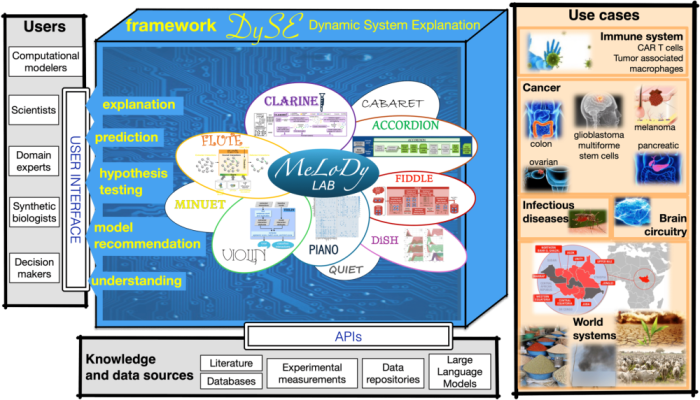
The amount of data and information produced by experimental laboratories studying complex biological systems is increasing at an incredible rate and is being disseminated at a growing number of publishing venues, scattering the knowledge and limiting the effectiveness and feasibility of manual analysis of all available information. This highlights the need for automated methods to retrieve and connect related pieces of the extensive yet fragmented collective knowledge, for the purpose of understanding, interpreting, explaining, and predicting the behavior of studied systems. To address this challenge, we have developed a tool suite, DySE. With DySE, we aim to integrate natural language processing, information retrieval, and artificial intelligence with assembly, inference, verification, and analysis of computational to enhance understanding and enable efficient interventions in complex systems. With DySE, we study the interplay between the immune system and diseases, including T cells, macrophages, glioblastoma multiforme stem cells, pancreatic cancer, melanoma, colon cancer, and ovarian cancer. DySE is featured in DARPA’s World Modelers program, where it assists decision-makers in identifying key points of intervention in complex world systems.
BioRECIPE
Biological system Representation for Evaluation, Curation, Interoperability, Preserving, and Execution
- A standardized biochemical interaction and executable model representation format that follows FAIR requirements
- [Paper][Documentation][Code]
FLUTE
The FiLter for Understanding True Events
- A filtering tool that utilizes existing databases to select biochemical interactions with high confidence
- [Paper][Documentation][Code]
CLARINET
CLARIfying NETworks
- A model recommendation tool that evaluates event collaboration graphs
- [Paper][Documentation][Code]
ACCORDION
ACCelerating and Optimizing model RecommenDatIONs
- A model recommendation tool that evaluates and selects relevant knowledge
- [Paper][Documentation] [Code]
VIOLIN
Validating Interactions Of Likely Importance to the Network
- A classification and scoring tool that evaluates biochemical interactions with respect to a model
- [Paper][Documentation] [Code]
FIDDLE
Finding Interactions using Diagram Driven modeL Extension
- A model extension tools that uses graph algorithms
- [Paper][Documentation][Code]
DiSH
Discrete Stochastic Heterogeneous simulator
- A simulator for hybrid models with a variety of simulation schemes and timing approaches
- [Paper][Documentation][Code]
CELESTA
Context Extraction through LEarning with Semi-supervised multi-Task Architecture
- [Paper][Documentation][Code]
Funding :
 |
 |
 |
 |
